Phospho-DRP1 (Ser637) Antibody - #DF2980
| Product: | Phospho-DRP1 (Ser637) Antibody |
| Catalog: | DF2980 |
| Description: | Rabbit polyclonal antibody to Phospho-DRP1 (Ser637) |
| Application: | WB IHC IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Horse, Sheep, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 82KD; 82kD(Calculated). |
| Uniprot: | O00429 |
| RRID: | AB_2840960 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF2980, RRID:AB_2840960.
Fold/Unfold
DLP1; dnm1l; DNM1L_HUMAN; Dnm1p/Vps1p-like protein; dnml1; DRP1; DVLP; Dymple; Dynamin 1 like; Dynamin family member proline-rich carboxyl-terminal domain less; Dynamin like protein; Dynamin related protein 1; Dynamin-1-like protein; Dynamin-like protein 4; Dynamin-like protein; Dynamin-like protein IV; Dynamin-related protein 1; DYNIV 11; EMPF; EMPF1; FLJ41912; HdynIV; VPS1;
Immunogens
Ubiquitously expressed with highest levels found in skeletal muscles, heart, kidney and brain. Isoform 1 is brain-specific. Isoform 2 and isoform 3 are predominantly expressed in testis and skeletal muscles respectively. Isoform 4 is weakly expressed in brain, heart and kidney. Isoform 5 is dominantly expressed in liver, heart and kidney. Isoform 6 is expressed in neurons.
- O00429 DNM1L_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MEALIPVINKLQDVFNTVGADIIQLPQIVVVGTQSSGKSSVLESLVGRDLLPRGTGIVTRRPLILQLVHVSQEDKRKTTGEENGVEAEEWGKFLHTKNKLYTDFDEIRQEIENETERISGNNKGVSPEPIHLKIFSPNVVNLTLVDLPGMTKVPVGDQPKDIELQIRELILRFISNPNSIILAVTAANTDMATSEALKISREVDPDGRRTLAVITKLDLMDAGTDAMDVLMGRVIPVKLGIIGVVNRSQLDINNKKSVTDSIRDEYAFLQKKYPSLANRNGTKYLARTLNRLLMHHIRDCLPELKTRINVLAAQYQSLLNSYGEPVDDKSATLLQLITKFATEYCNTIEGTAKYIETSELCGGARICYIFHETFGRTLESVDPLGGLNTIDILTAIRNATGPRPALFVPEVSFELLVKRQIKRLEEPSLRCVELVHEEMQRIIQHCSNYSTQELLRFPKLHDAIVEVVTCLLRKRLPVTNEMVHNLVAIELAYINTKHPDFADACGLMNNNIEEQRRNRLARELPSAVSRDKSSKVPSALAPASQEPSPAASAEADGKLIQDSRRETKNVASGGGGVGDGVQEPTTGNWRGMLKTSKAEELLAEEKSKPIPIMPASPQKGHAVNLLDVPVPVARKLSAREQRDCEVIERLIKSYFLIVRKNIQDSVPKAVMHFLVNHVKDTLQSELVGQLYKSSLLDDLLTESEDMAQRRKEAADMLKALQGASQIIAEIRETHLW
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
PTMs - O00429 As Substrate
| Site | PTM Type | Enzyme | Source |
|---|---|---|---|
| M1 | Acetylation | Uniprot | |
| S40 | Phosphorylation | P49841 (GSK3B) | Uniprot |
| S44 | Phosphorylation | P49841 (GSK3B) | Uniprot |
| T78 | Phosphorylation | Uniprot | |
| T79 | Phosphorylation | Uniprot | |
| K92 | Ubiquitination | Uniprot | |
| K97 | Ubiquitination | Uniprot | |
| K99 | Ubiquitination | Uniprot | |
| T102 | Phosphorylation | Uniprot | |
| R108 | Methylation | Uniprot | |
| K123 | Ubiquitination | Uniprot | |
| S126 | Phosphorylation | Uniprot | |
| K133 | Ubiquitination | Uniprot | |
| S136 | Phosphorylation | Uniprot | |
| K160 | Ubiquitination | Uniprot | |
| S179 | Phosphorylation | Uniprot | |
| T193 | Phosphorylation | Uniprot | |
| S200 | Phosphorylation | Uniprot | |
| K238 | Ubiquitination | Uniprot | |
| K255 | Ubiquitination | Uniprot | |
| K256 | Ubiquitination | Uniprot | |
| Y266 | Phosphorylation | Uniprot | |
| K271 | Ubiquitination | Uniprot | |
| K272 | Ubiquitination | Uniprot | |
| K283 | Acetylation | Uniprot | |
| S330 | Phosphorylation | Uniprot | |
| C361 | S-Nitrosylation | Uniprot | |
| C367 | S-Nitrosylation | Uniprot | |
| Y368 | Phosphorylation | Uniprot | |
| T394 | Phosphorylation | Uniprot | |
| T400 | Phosphorylation | Uniprot | |
| S412 | Phosphorylation | Uniprot | |
| Y449 | Phosphorylation | Uniprot | |
| S529 | Phosphorylation | Uniprot | |
| K532 | Sumoylation | Uniprot | |
| K535 | Sumoylation | Uniprot | |
| K535 | Ubiquitination | Uniprot | |
| S544 | Phosphorylation | Uniprot | |
| S548 | Phosphorylation | Uniprot | |
| S552 | Phosphorylation | Uniprot | |
| K558 | Sumoylation | Uniprot | |
| K568 | Sumoylation | Uniprot | |
| K568 | Ubiquitination | Uniprot | |
| S572 | Phosphorylation | Uniprot | |
| T586 | Phosphorylation | Uniprot | |
| K594 | Sumoylation | Uniprot | |
| T595 | Phosphorylation | Uniprot | |
| K597 | Sumoylation | Uniprot | |
| K597 | Ubiquitination | Uniprot | |
| K606 | Sumoylation | Uniprot | |
| S607 | Phosphorylation | Uniprot | |
| K608 | Sumoylation | Uniprot | |
| S616 | Phosphorylation | P06493 (CDK1) , P28482 (MAPK1) , P27361 (MAPK3) , Q05655 (PRKCD) , Q00535 (CDK5) , P24941 (CDK2) | Uniprot |
| S637 | Phosphorylation | P17612 (PRKACA) , Q13464 (ROCK1) , Q13131 (PRKAA1) | Uniprot |
| C644 | S-Nitrosylation | Uniprot | |
| S693 | Phosphorylation | P49841 (GSK3B) | Uniprot |
| T701 | Phosphorylation | Uniprot | |
| S724 | Phosphorylation | Uniprot |
Research Backgrounds
Functions in mitochondrial and peroxisomal division. Mediates membrane fission through oligomerization into membrane-associated tubular structures that wrap around the scission site to constrict and sever the mitochondrial membrane through a GTP hydrolysis-dependent mechanism. The specific recruitment at scission sites is mediated by membrane receptors like MFF, MIEF1 and MIEF2 for mitochondrial membranes. While the recruitment by the membrane receptors is GTP-dependent, the following hydrolysis of GTP induces the dissociation from the receptors and allows DNM1L filaments to curl into closed rings that are probably sufficient to sever a double membrane. Through its function in mitochondrial division, ensures the survival of at least some types of postmitotic neurons, including Purkinje cells, by suppressing oxidative damage. Required for normal brain development, including that of cerebellum. Facilitates developmentally regulated apoptosis during neural tube formation. Required for a normal rate of cytochrome c release and caspase activation during apoptosis; this requirement may depend upon the cell type and the physiological apoptotic cues. Plays an important role in mitochondrial fission during mitosis. Required for formation of endocytic vesicles. Proposed to regulate synaptic vesicle membrane dynamics through association with BCL2L1 isoform Bcl-X(L) which stimulates its GTPase activity in synaptic vesicles; the function may require its recruitment by MFF to clathrin-containing vesicles. Required for programmed necrosis execution. Rhythmic control of its activity following phosphorylation at Ser-637 is essential for the circadian control of mitochondrial ATP production.
Inhibits peroxisomal division when overexpressed.
Inhibits peroxisomal division when overexpressed.
Phosphorylation/dephosphorylation events on two sites near the GED domain regulate mitochondrial fission. Phosphorylation on Ser-637 inhibits the GTPase activity, leading to a defect in mitochondrial fission promoting mitochondrial elongation. Dephosphorylated on this site by PPP3CA which promotes mitochondrial fission. Phosphorylation on Ser-616 activates the GTPase activity and promotes mitochondrial fission. Phosphorylated in a circadian manner at Ser-637.
Sumoylated on various lysine residues within the B domain, probably by MUL1. Sumoylation positively regulates mitochondrial fission. Desumoylated by SENP5 during G2/M transition of mitosis. Appears to be linked to its catalytic activity.
S-nitrosylation increases DNM1L dimerization, mitochondrial fission and causes neuronal damage.
Ubiquitination by MARCHF5 affects mitochondrial morphology.
O-GlcNAcylation augments the level of the GTP-bound active form of DRP1 and induces translocation from the cytoplasm to mitochondria in cardiomyocytes. It also decreases phosphorylation at Ser-637 (By similarity).
Cytoplasm>Cytosol. Golgi apparatus. Endomembrane system>Peripheral membrane protein. Mitochondrion outer membrane>Peripheral membrane protein. Peroxisome. Membrane>Clathrin-coated pit. Cytoplasmic vesicle>Secretory vesicle>Synaptic vesicle membrane.
Note: Mainly cytosolic. Translocated to the mitochondrial membrane through O-GlcNAcylation and interaction with FIS1. Recruited to the mitochondrial outer membrane by interaction with MIEF1. Colocalized with MARCHF5 at mitochondrial membrane. Localizes to mitochondria at sites of division. Localizes to mitochondria following necrosis induction. Associated with peroxisomal membranes, partly recruited there by PEX11B. May also be associated with endoplasmic reticulum tubules and cytoplasmic vesicles and found to be perinuclear. In some cell types, localizes to the Golgi complex. Binds to phospholipid membranes.
Ubiquitously expressed with highest levels found in skeletal muscles, heart, kidney and brain. Isoform 1 is brain-specific. Isoform 2 and isoform 3 are predominantly expressed in testis and skeletal muscles respectively. Isoform 4 is weakly expressed in brain, heart and kidney. Isoform 5 is dominantly expressed in liver, heart and kidney. Isoform 6 is expressed in neurons.
Homotetramer; dimerizes through the N-terminal GTP-middle region of one molecule binding to the GED domain of another DNM1L molecule. Oligomerizes in a GTP-dependent manner to form membrane-associated tubules with a spiral pattern. Interacts with GSK3B and MARCHF5. Interacts (via the GTPase and B domains) with UBE2I; the interaction promotes sumoylation of DNM1L, mainly in its B domain. Interacts with PPP3CA; the interaction dephosphorylates DNM1L and regulates its transition to mitochondria. Interacts with BCL2L1 isoform BCL-X(L) and CLTA; DNM1L and BCL2L1 isoform BCL-X(L) may form a complex in synaptic vesicles that also contains clathrin and MFF. Interacts with FIS1. Interacts with MIEF2 and MIEF1; GTP-dependent this regulates GTP hydrolysis and DNM1L oligomerization. Interacts with PGAM5; this interaction leads to dephosphorylation at Ser-656 and activation of GTPase activity and eventually to mitochondria fragmentation.
The GED domain folds back to interact, in cis, with the GTP-binding domain and middle domain, and interacts, in trans, with the GED domains of other DNM1L molecules, and is thus critical for activating GTPase activity and for DNM1L dimerization.
Belongs to the TRAFAC class dynamin-like GTPase superfamily. Dynamin/Fzo/YdjA family.
Research Fields
· Cellular Processes > Cell growth and death > Necroptosis. (View pathway)
· Environmental Information Processing > Signal transduction > TNF signaling pathway. (View pathway)
· Organismal Systems > Immune system > NOD-like receptor signaling pathway. (View pathway)
References
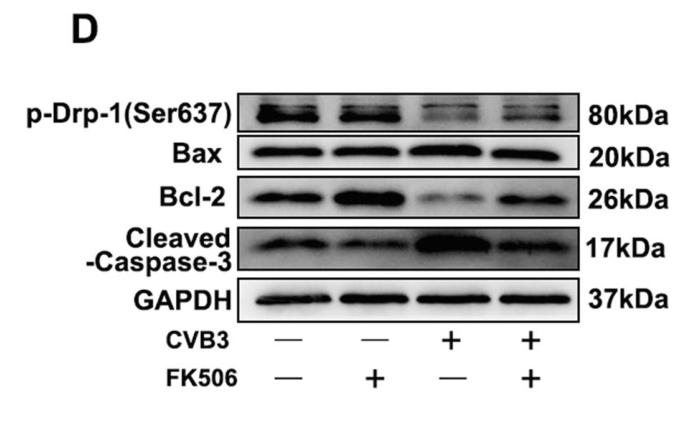
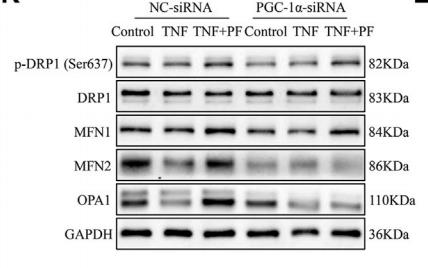
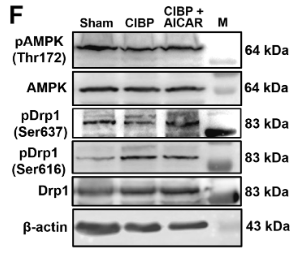
Application: WB Species: Rat Sample: brain microvascular endothelial cells
Application: WB Species: Mice Sample: NRCMs
Application: WB Species: rat Sample: C2C12 cells
Application: IF/ICC Species: mouse Sample: oocytes
Application: WB Species: Rat Sample: spinal cord
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.