ASPM Antibody - #DF10064
| Product: | ASPM Antibody |
| Catalog: | DF10064 |
| Description: | Rabbit polyclonal antibody to ASPM |
| Application: | WB IHC IF/ICC |
| Reactivity: | Human, Rat |
| Prediction: | Bovine, Horse, Sheep, Dog |
| Mol.Wt.: | 410 kDa; 410kD(Calculated). |
| Uniprot: | Q8IZT6 |
| RRID: | AB_2840644 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF10064, RRID:AB_2840644.
Immunogens
- Q8IZT6 ASPM_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MANRRVGRGCWEVSPTERRPPAGLRGPAAEEEASSPPVLSLSHFCRSPFLCFGDVLLGASRTLSLALDNPNEEVAEVKISHFPAADLGFSVSQRCFVLQPKEKIVISVNWTPLKEGRVREIMTFLVNDVLKHQAILLGNAEEQKKKKRSLWDTIKKKKISASTSHNRRVSNIQNVNKTFSVSQKVDRVRSPLQACENLAMNEGGPPTENNSLILEENKIPISPISPAFNECHGATCLPLSVRRSTTYSSLHASENRELLNVHSANVSKVSFNEKAVTETSFNSVNVNGQRGENSKLSLTPNCSSTLNITQSQIHFLSPDSFVNNSHGANNELELVTCLSSDMFMKDNSQPVHLESTIAHEIYQKILSPDSFIKDNYGLNQDLESESVNPILSPNQFLKDNMAYMCTSQQTCKVPLSNENSQVPQSPEDWRKSEVSPRIPECQGSKSPKAIFEELVEMKSNYYSFIKQNNPKFSAVQDISSHSHNKQPKRRPILSATVTKRKATCTRENQTEINKPKAKRCLNSAVGEHEKVINNQKEKEDFHSYLPIIDPILSKSKSYKNEVTPSSTTASVARKRKSDGSMEDANVRVAITEHTEVREIKRIHFSPSEPKTSAVKKTKNVTTPISKRISNREKLNLKKKTDLSIFRTPISKTNKRTKPIIAVAQSSLTFIKPLKTDIPRHPMPFAAKNMFYDERWKEKQEQGFTWWLNFILTPDDFTVKTNISEVNAATLLLGIENQHKISVPRAPTKEEMSLRAYTARCRLNRLRRAACRLFTSEKMVKAIKKLEIEIEARRLIVRKDRHLWKDVGERQKVLNWLLSYNPLWLRIGLETTYGELISLEDNSDVTGLAMFILNRLLWNPDIAAEYRHPTVPHLYRDGHEEALSKFTLKKLLLLVCFLDYAKISRLIDHDPCLFCKDAEFKASKEILLAFSRDFLSGEGDLSRHLGLLGLPVNHVQTPFDEFDFAVTNLAVDLQCGVRLVRTMELLTQNWDLSKKLRIPAISRLQKMHNVDIVLQVLKSRGIELSDEHGNTILSKDIVDRHREKTLRLLWKIAFAFQVDISLNLDQLKEEIAFLKHTKSIKKTISLLSCHSDDLINKKKGKRDSGSFEQYSENIKLLMDWVNAVCAFYNKKVENFTVSFSDGRVLCYLIHHYHPCYVPFDAICQRTTQTVECTQTGSVVLNSSSESDDSSLDMSLKAFDHENTSELYKELLENEKKNFHLVRSAVRDLGGIPAMINHSDMSNTIPDEKVVITYLSFLCARLLDLRKEIRAARLIQTTWRKYKLKTDLKRHQEREKAARIIQLAVINFLAKQRLRKRVNAALVIQKYWRRVLAQRKLLMLKKEKLEKVQNKAASLIQGYWRRYSTRQRFLKLKYYSIILQSRIRMIIAVTSYKRYLWATVTIQRHWRAYLRRKQDQQRYEMLKSSTLIIQSMFRKWKQRKMQSQVKATVILQRAFREWHLRKQAKEENSAIIIQSWYRMHKELRKYIYIRSCVVIIQKRFRCFQAQKLYKRRKESILTIQKYYKAYLKGKIERTNYLQKRAAAIQLQAAFRRLKAHNLCRQIRAACVIQSYWRMRQDRVRFLNLKKTIIKFQAHVRKHQQRQKYKKMKKAAVIIQTHFRAYIFAMKVLASYQKTRSAVIVLQSAYRGMQARKMYIHILTSVIKIQSYYRAYVSKKEFLSLKNATIKLQSTVKMKQTRKQYLHLRAAALFIQQCYRSKKIAAQKREEYMQMRESCIKLQAFVRGYLVRKQMRLQRKAVISLQSYFRMRKARQYYLKMYKAIIVIQNYYHAYKAQVNQRKNFLQVKKAATCLQAAYRGYKVRQLIKQQSIAALKIQSAFRGYNKRVKYQSVLQSIIKIQRWYRAYKTLHDTRTHFLKTKAAVISLQSAYRGWKVRKQIRREHQAALKIQSAFRMAKAQKQFRLFKTAALVIQQNFRAWTAGRKQCMEYIELRHAVLVLQSMWKGKTLRRQLQRQHKCAIIIQSYYRMHVQQKKWKIMKKAALLIQKYYRAYSIGREQNHLYLKTKAAVVTLQSAYRGMKVRKRIKDCNKAAVTIQSKYRAYKTKKKYATYRASAIIIQRWYRGIKITNHQHKEYLNLKKTAIKIQSVYRGIRVRRHIQHMHRAATFIKAMFKMHQSRISYHTMRKAAIVIQVRCRAYYQGKMQREKYLTILKAVKVLQASFRGVRVRRTLRKMQTAATLIQSNYRRYRQQTYFNKLKKITKTVQQRYWAMKERNIQFQRYNKLRHSVIYIQAIFRGKKARRHLKMMHIAATLIQRRFRTLMMRRRFLSLKKTAILIQRKYRAHLCTKHHLQFLQVQNAVIKIQSSYRRWMIRKRMREMHRAATFIQSTFRMHRLHMRYQALKQASVVIQQQYQANRAAKLQRQHYLRQRHSAVILQAAFRGMKTRRHLKSMHSSATLIQSRFRSLLVRRRFISLKKATIFVQRKYRATICAKHKLYQFLHLRKAAITIQSSYRRLMVKKKLQEMQRAAVLIQATFRMYRTYITFQTWKHASILIQQHYRTYRAAKLQRENYIRQWHSAVVIQAAYKGMKARQLLREKHKASIVIQSTYRMYRQYCFYQKLQWATKIIQEKYRANKKKQKVFQHNELKKETCVQAGFQDMNIKKQIQEQHQAAIIIQKHCKAFKIRKHYLHLRATVVSIQRRYRKLTAVRTQAVICIQSYYRGFKVRKDIQNMHRAATLIQSFYRMHRAKVDYETKKTAIVVIQNYYRLYVRVKTERKNFLAVQKSVRTIQAAFRGMKVRQKLKNVSEEKMAAIVNQSALCCYRSKTQYEAVQSEGVMIQEWYKASGLACSQEAEYHSQSRAAVTIQKAFCRMVTRKLETQKCAALRIQFFLQMAVYRRRFVQQKRAAITLQHYFRTWQTRKQFLLYRKAAVVLQNHYRAFLSAKHQRQVYLQIRSSVIIIQARSKGFIQKRKFQEIKNSTIKIQAMWRRYRAKKYLCKVKAACKIQAWYRCWRAHKEYLAILKAVKIIQGCFYTKLERTRFLNVRASAIIIQRKWRAILPAKIAHEHFLMIKRHRAACLIQAHYRGYKGRQVFLRQKSAALIIQKYIRAREAGKHERIKYIEFKKSTVILQALVRGWLVRKRFLEQRAKIRLLHFTAAAYYHLNAVRIQRAYKLYLAVKNANKQVNSVICIQRWFRARLQEKRFIQKYHSIKKIEHEGQECLSQRNRAASVIQKAVRHFLLRKKQEKFTSGIIKIQALWRGYSWRKKNDCTKIKAIRLSLQVVNREIREENKLYKRTALALHYLLTYKHLSAILEALKHLEVVTRLSPLCCENMAQSGAISKIFVLIRSCNRSIPCMEVIRYAVQVLLNVSKYEKTTSAVYDVENCIDILLELLQIYREKPGNKVADKGGSIFTKTCCLLAILLKTTNRASDVRSRSKVVDRIYSLYKLTAHKHKMNTERILYKQKKNSSISIPFIPETPVRTRIVSRLKPDWVLRRDNMEEITNPLQAIQMVMDTLGIPY
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
PTMs - Q8IZT6 As Substrate
| Site | PTM Type | Enzyme | Source |
|---|---|---|---|
| R8 | Methylation | Uniprot | |
| S14 | Phosphorylation | Uniprot | |
| S35 | Phosphorylation | Uniprot | |
| K101 | Ubiquitination | Uniprot | |
| T111 | Phosphorylation | Uniprot | |
| K144 | Sumoylation | Uniprot | |
| K144 | Ubiquitination | Uniprot | |
| T153 | Phosphorylation | Uniprot | |
| S170 | Phosphorylation | Uniprot | |
| K177 | Ubiquitination | Uniprot | |
| T178 | Phosphorylation | Uniprot | |
| S180 | Phosphorylation | Uniprot | |
| K184 | Sumoylation | Uniprot | |
| K184 | Ubiquitination | Uniprot | |
| S190 | Phosphorylation | Uniprot | |
| S211 | Phosphorylation | Uniprot | |
| S222 | Phosphorylation | Uniprot | |
| S225 | Phosphorylation | Uniprot | |
| T245 | Phosphorylation | Uniprot | |
| T246 | Phosphorylation | Uniprot | |
| Y247 | Phosphorylation | Uniprot | |
| S249 | Phosphorylation | Uniprot | |
| S263 | Phosphorylation | Uniprot | |
| S267 | Phosphorylation | Uniprot | |
| S270 | Phosphorylation | Uniprot | |
| K274 | Ubiquitination | Uniprot | |
| T279 | Phosphorylation | Uniprot | |
| S280 | Phosphorylation | Uniprot | |
| S283 | Phosphorylation | Uniprot | |
| S297 | Phosphorylation | Uniprot | |
| S348 | Phosphorylation | Uniprot | |
| S355 | Phosphorylation | Uniprot | |
| Y362 | Phosphorylation | Uniprot | |
| K364 | Ubiquitination | Uniprot | |
| S367 | Phosphorylation | Uniprot | |
| S370 | Phosphorylation | Uniprot | |
| K373 | Ubiquitination | Uniprot | |
| S392 | Phosphorylation | Uniprot | |
| K398 | Ubiquitination | Uniprot | |
| S407 | Phosphorylation | Uniprot | |
| S416 | Phosphorylation | Uniprot | |
| S420 | Phosphorylation | Uniprot | |
| S425 | Phosphorylation | Uniprot | |
| K431 | Ubiquitination | Uniprot | |
| S432 | Phosphorylation | Uniprot | |
| S435 | Phosphorylation | Uniprot | |
| S444 | Phosphorylation | Uniprot | |
| K445 | Ubiquitination | Uniprot | |
| S446 | Phosphorylation | Uniprot | |
| Y461 | Phosphorylation | Uniprot | |
| Y462 | Phosphorylation | Uniprot | |
| S463 | Phosphorylation | Uniprot | |
| K466 | Ubiquitination | Uniprot | |
| K471 | Ubiquitination | Uniprot | |
| S473 | Phosphorylation | Uniprot | |
| S482 | Phosphorylation | Uniprot | |
| K485 | Ubiquitination | Uniprot | |
| S494 | Phosphorylation | Uniprot | |
| T496 | Phosphorylation | Uniprot | |
| K514 | Ubiquitination | Uniprot | |
| K530 | Acetylation | Uniprot | |
| K530 | Ubiquitination | Uniprot | |
| K538 | Ubiquitination | Uniprot | |
| S553 | Phosphorylation | Uniprot | |
| K554 | Ubiquitination | Uniprot | |
| K559 | Sumoylation | Uniprot | |
| K559 | Ubiquitination | Uniprot | |
| T563 | Phosphorylation | Uniprot | |
| S565 | Phosphorylation | Uniprot | |
| S570 | Phosphorylation | Uniprot | |
| S577 | Phosphorylation | Uniprot | |
| S580 | Phosphorylation | Uniprot | |
| S605 | Phosphorylation | Uniprot | |
| S607 | Phosphorylation | Uniprot | |
| K610 | Ubiquitination | Uniprot | |
| T611 | Phosphorylation | Uniprot | |
| S612 | Phosphorylation | Uniprot | |
| K615 | Acetylation | Uniprot | |
| K616 | Acetylation | Uniprot | |
| K618 | Ubiquitination | Uniprot | |
| T621 | Phosphorylation | Uniprot | |
| T622 | Phosphorylation | Uniprot | |
| S625 | Phosphorylation | Uniprot | |
| K626 | Ubiquitination | Uniprot | |
| K639 | Ubiquitination | Uniprot | |
| T640 | Phosphorylation | Uniprot | |
| S643 | Phosphorylation | Uniprot | |
| T647 | Phosphorylation | Uniprot | |
| S650 | Phosphorylation | Uniprot | |
| K651 | Ubiquitination | Uniprot | |
| K674 | Ubiquitination | Uniprot | |
| K687 | Ubiquitination | Uniprot | |
| K696 | Ubiquitination | Uniprot | |
| T729 | Phosphorylation | Uniprot | |
| K739 | Ubiquitination | Uniprot | |
| T747 | Phosphorylation | Uniprot | |
| K748 | Ubiquitination | Uniprot | |
| S752 | Phosphorylation | Uniprot | |
| K777 | Ubiquitination | Uniprot | |
| K804 | Ubiquitination | Uniprot | |
| K915 | Ubiquitination | Uniprot | |
| K923 | Ubiquitination | Uniprot | |
| K1017 | Ubiquitination | Uniprot | |
| S1018 | Phosphorylation | Uniprot | |
| S1024 | Phosphorylation | Uniprot | |
| T1030 | Phosphorylation | Uniprot | |
| K1034 | Ubiquitination | Uniprot | |
| K1074 | Ubiquitination | Uniprot | |
| K1081 | Ubiquitination | Uniprot | |
| S1084 | Phosphorylation | Uniprot | |
| S1090 | Phosphorylation | Uniprot | |
| K1096 | Ubiquitination | Uniprot | |
| R1101 | Methylation | Uniprot | |
| S1103 | Phosphorylation | Uniprot | |
| S1105 | Phosphorylation | Uniprot | |
| S1110 | Phosphorylation | Uniprot | |
| K1207 | Ubiquitination | Uniprot | |
| K1215 | Ubiquitination | Uniprot | |
| K1279 | Acetylation | Uniprot | |
| Y1325 | Phosphorylation | Uniprot | |
| K1334 | Ubiquitination | Uniprot | |
| K1339 | Acetylation | Uniprot | |
| K1340 | Acetylation | Uniprot | |
| K1345 | Ubiquitination | Uniprot | |
| K1349 | Ubiquitination | Uniprot | |
| Y1361 | Phosphorylation | Uniprot | |
| Y1417 | Phosphorylation | Uniprot | |
| Y1484 | Phosphorylation | Uniprot | |
| K1511 | Ubiquitination | Uniprot | |
| S1513 | Phosphorylation | Uniprot | |
| T1516 | Phosphorylation | Uniprot | |
| K1519 | Ubiquitination | Uniprot | |
| K1522 | Ubiquitination | Uniprot | |
| K1537 | Ubiquitination | Uniprot | |
| K1552 | Ubiquitination | Uniprot | |
| S1641 | Phosphorylation | Uniprot | |
| Y1643 | Phosphorylation | Uniprot | |
| S1658 | Phosphorylation | Uniprot | |
| K1679 | Ubiquitination | Uniprot | |
| K1684 | Ubiquitination | Uniprot | |
| K1696 | Ubiquitination | Uniprot | |
| Y1770 | Phosphorylation | Uniprot | |
| K1796 | Ubiquitination | Uniprot | |
| K1802 | Ubiquitination | Uniprot | |
| K1803 | Ubiquitination | Uniprot | |
| K1822 | Ubiquitination | Uniprot | |
| S1825 | Phosphorylation | Uniprot | |
| K1830 | Ubiquitination | Uniprot | |
| S1833 | Phosphorylation | Uniprot | |
| K1843 | Ubiquitination | Uniprot | |
| Y1844 | Phosphorylation | Uniprot | |
| S1846 | Phosphorylation | Uniprot | |
| S1850 | Phosphorylation | Uniprot | |
| K1862 | Ubiquitination | Uniprot | |
| K1873 | Ubiquitination | Uniprot | |
| K1875 | Ubiquitination | Uniprot | |
| K1903 | Acetylation | Uniprot | |
| K1903 | Ubiquitination | Uniprot | |
| S1906 | Phosphorylation | Uniprot | |
| K1912 | Acetylation | Uniprot | |
| K1915 | Acetylation | Uniprot | |
| K1939 | Ubiquitination | Uniprot | |
| Y1944 | Phosphorylation | Uniprot | |
| K1972 | Ubiquitination | Uniprot | |
| K1995 | Ubiquitination | Uniprot | |
| K2002 | Ubiquitination | Uniprot | |
| K2021 | Ubiquitination | Uniprot | |
| S2029 | Phosphorylation | Uniprot | |
| K2045 | Ubiquitination | Uniprot | |
| K2053 | Acetylation | Uniprot | |
| K2053 | Ubiquitination | Uniprot | |
| Y2063 | Phosphorylation | Uniprot | |
| S2069 | Phosphorylation | Uniprot | |
| K2088 | Ubiquitination | Uniprot | |
| K2094 | Ubiquitination | Uniprot | |
| T2096 | Phosphorylation | Uniprot | |
| K2099 | Ubiquitination | Uniprot | |
| S2102 | Phosphorylation | Uniprot | |
| Y2104 | Phosphorylation | Uniprot | |
| T2121 | Phosphorylation | Uniprot | |
| K2124 | Ubiquitination | Uniprot | |
| S2132 | Phosphorylation | Uniprot | |
| S2135 | Phosphorylation | Uniprot | |
| Y2136 | Phosphorylation | Uniprot | |
| T2138 | Phosphorylation | Uniprot | |
| K2157 | Ubiquitination | Uniprot | |
| K2162 | Ubiquitination | Uniprot | |
| K2168 | Ubiquitination | Uniprot | |
| K2171 | Ubiquitination | Uniprot | |
| S2176 | Phosphorylation | Uniprot | |
| K2188 | Ubiquitination | Uniprot | |
| T2191 | Phosphorylation | Uniprot | |
| K2217 | Ubiquitination | Uniprot | |
| K2227 | Ubiquitination | Uniprot | |
| K2260 | Methylation | Uniprot | |
| K2287 | Ubiquitination | Uniprot | |
| K2295 | Acetylation | Uniprot | |
| K2358 | Ubiquitination | Uniprot | |
| K2405 | Ubiquitination | Uniprot | |
| S2429 | Phosphorylation | Uniprot | |
| K2432 | Ubiquitination | Uniprot | |
| K2459 | Ubiquitination | Uniprot | |
| T2490 | Phosphorylation | Uniprot | |
| S2557 | Phosphorylation | Uniprot | |
| Y2570 | Phosphorylation | Uniprot | |
| Y2573 | Phosphorylation | Uniprot | |
| K2581 | Ubiquitination | Uniprot | |
| K2586 | Ubiquitination | Uniprot | |
| K2595 | Ubiquitination | Uniprot | |
| K2603 | Ubiquitination | Uniprot | |
| K2604 | Ubiquitination | Uniprot | |
| K2618 | Ubiquitination | Uniprot | |
| K2619 | Ubiquitination | Uniprot | |
| K2633 | Ubiquitination | Uniprot | |
| Y2675 | Phosphorylation | Uniprot | |
| Y2676 | Phosphorylation | Uniprot | |
| T2730 | Phosphorylation | Uniprot | |
| K2733 | Ubiquitination | Uniprot | |
| K2740 | Ubiquitination | Uniprot | |
| K2759 | Ubiquitination | Uniprot | |
| K2765 | Ubiquitination | Uniprot | |
| K2781 | Ubiquitination | Uniprot | |
| S2806 | Phosphorylation | Uniprot | |
| K2823 | Ubiquitination | Uniprot | |
| K2837 | Ubiquitination | Uniprot | |
| K2877 | Ubiquitination | Uniprot | |
| K2884 | Ubiquitination | Uniprot | |
| S2912 | Phosphorylation | Uniprot | |
| K2933 | Ubiquitination | Uniprot | |
| K2960 | Ubiquitination | Uniprot | |
| Y2974 | Phosphorylation | Uniprot | |
| K2991 | Ubiquitination | Uniprot | |
| S3003 | Phosphorylation | Uniprot | |
| K3018 | Ubiquitination | Uniprot | |
| K3028 | Ubiquitination | Uniprot | |
| K3053 | Ubiquitination | Uniprot | |
| S3054 | Phosphorylation | Uniprot | |
| K3061 | Ubiquitination | Uniprot | |
| S3082 | Phosphorylation | Uniprot | |
| K3135 | Ubiquitination | Uniprot | |
| K3139 | Ubiquitination | Uniprot | |
| K3163 | Ubiquitination | Uniprot | |
| K3169 | Ubiquitination | Uniprot | |
| S3186 | Phosphorylation | Uniprot | |
| K3190 | Ubiquitination | Uniprot | |
| K3203 | Ubiquitination | Uniprot | |
| S3235 | Phosphorylation | Uniprot | |
| K3364 | Ubiquitination | Uniprot | |
| S3387 | Phosphorylation | Uniprot | |
| Y3400 | Phosphorylation | Uniprot | |
| K3404 | Ubiquitination | Uniprot | |
| T3406 | Phosphorylation | Uniprot | |
| K3420 | Acetylation | Uniprot | |
| K3423 | Acetylation | Uniprot | |
| K3423 | Ubiquitination | Uniprot | |
| S3425 | Phosphorylation | Uniprot | |
| T3435 | Phosphorylation | Uniprot | |
| T3439 | Phosphorylation | Uniprot | |
| S3443 | Phosphorylation | Uniprot | |
| K3446 | Ubiquitination | Uniprot | |
| Y3477 | Phosphorylation | Uniprot |
Research Backgrounds
Involved in mitotic spindle regulation and coordination of mitotic processes. The function in regulating microtubule dynamics at spindle poles including spindle orientation, astral microtubule density and poleward microtubule flux seems to depend on the association with the katanin complex formed by KATNA1 and KATNB1. Enhances the microtubule lattice severing activity of KATNA1 by recruiting the katanin complex to microtubules. Can block microtubule minus-end growth and reversely this function can be enhanced by the katanin complex. May have a preferential role in regulating neurogenesis.
Cytoplasm. Cytoplasm>Cytoskeleton>Spindle. Nucleus.
Note: The nuclear-cytoplasmic distribution could be regulated by the availability of calmodulin (By similarity). Localizes to spindle poles during mitosis (PubMed:19690332). Associates with microtubule minus ends (By similarity).
Interacts with KATNA1 and KATNB1; katanin complex formation KATNA1:KATNB1 is required for the association.
References
Application: IHC Species: Human Sample: KIRC and LIHC tissues
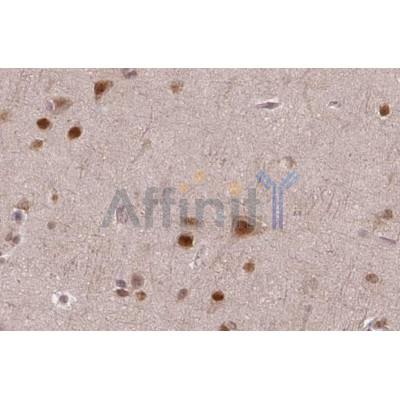
Application: IHC Species: Rat Sample:
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.